Abstract
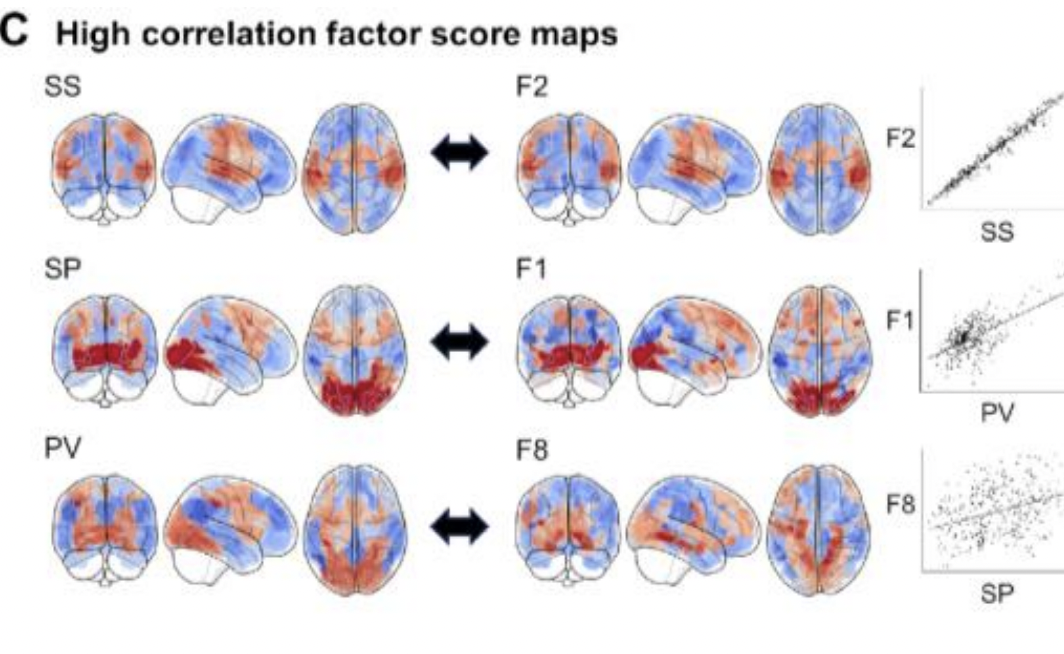
Despite the widespread use of the Research Domain Criteria (RDoC) framework in psychiatry and neuroscience, recent studies suggest that the RDoC is insufficiently specific or excessively broad relative to the underlying brain circuitry it seeks to elucidate. To address these concerns, we employ a latent variable approach using bifactor analysis. We examine 84 whole-brain task-based fMRI (tfMRI) activation maps from 19 studies with 6192 participants. A curated subset of 37 maps with a balanced representation of RDoC domains constitute the training set, and the remaining held-out maps form the internal validation set. External validation is conducted using 36 peak coordinate activation maps from Neurosynth, using terms of RDoC constructs as seeds for topic meta-analysis. Here, we show that a bifactor model incorporating a task-general domain and splitting the cognitive systems domain better fits the examined corpus of tfMRI data than the current RDoC framework. We also identify the domain of arousal and regulatory systems as underrepresented. Our data-driven validation supports revising the RDoC framework to reflect underlying brain circuitry more accurately.