Abstract
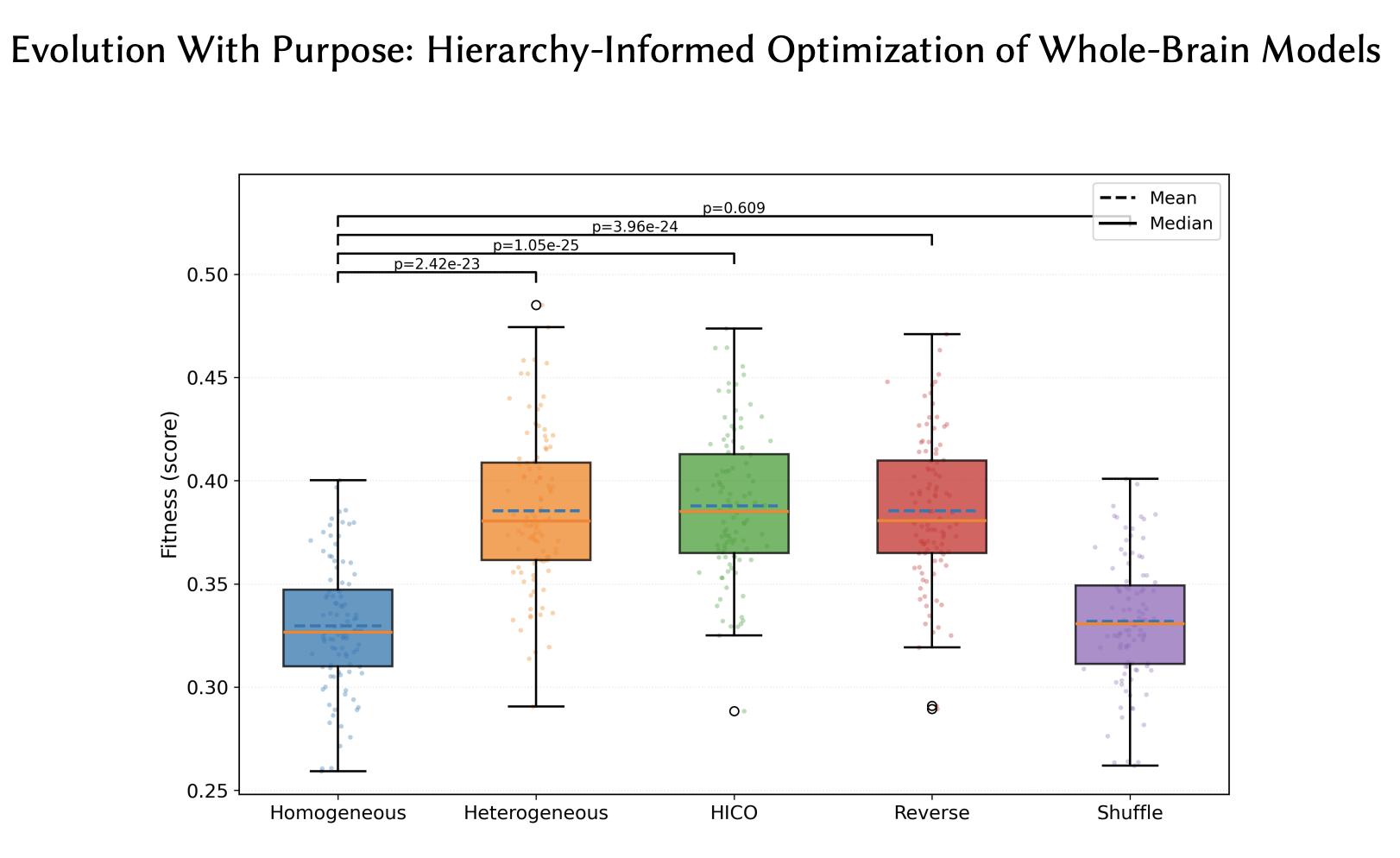
Evolutionary search is well suited for large-scale biophysical brain modeling, where many parameters with nonlinear interactions and no tractable gradients need to be optimized. Standard evolutionary approaches achieve an excellent fit to MRI data; however, among many possible such solutions, it finds ones that overfit to individual subjects and provide limited predictive power. This paper investigates whether guiding evolution with biological knowledge can help. Focusing on whole-brain Dynamic Mean Field (DMF) models, a baseline where 20 parameters were shared across the brain was compared against a heterogeneous formulation where different sets of 20 parameters were used for the seven canonical brain regions. The heterogeneous model was optimized using four strategies: optimizing all parameters at once, a curricular approach following the hierarchy of brain networks (HICO), a reversed curricular approach, and a randomly shuffled curricular approach. While all heterogeneous strategies fit the data well, only curricular approaches generalized to new subjects. Most importantly, only HICO made it possible to use the parameter sets to predict the subjects’ behavioral abilities as well. Thus, by guiding evolution with biological knowledge about the hierarchy of brain regions, HICO demonstrated how domain knowledge can be harnessed to serve the purpose of optimization in real-world domains.